Function that performs Principal Coordinates Analysis on abundance matrix.
PCO(x, dim, distfun) # S3 method for default PCO(x, dim = 3) # S3 method for Dataset PCO(x, dim = 3, distfun = dist)
Arguments
| x | Distance matrix, must be a |
|---|---|
| dim | Number of dimensions to return. |
| distfun | Function that calculates distance matrix for abundance
table in |
Value
A PCO object as defined by the pco function.
Details
This is the same as function pco from the labdsv,
but it includes a Dataset method.
See also
create_dataset, pca, PCA,
pco, dsvdis
Examples
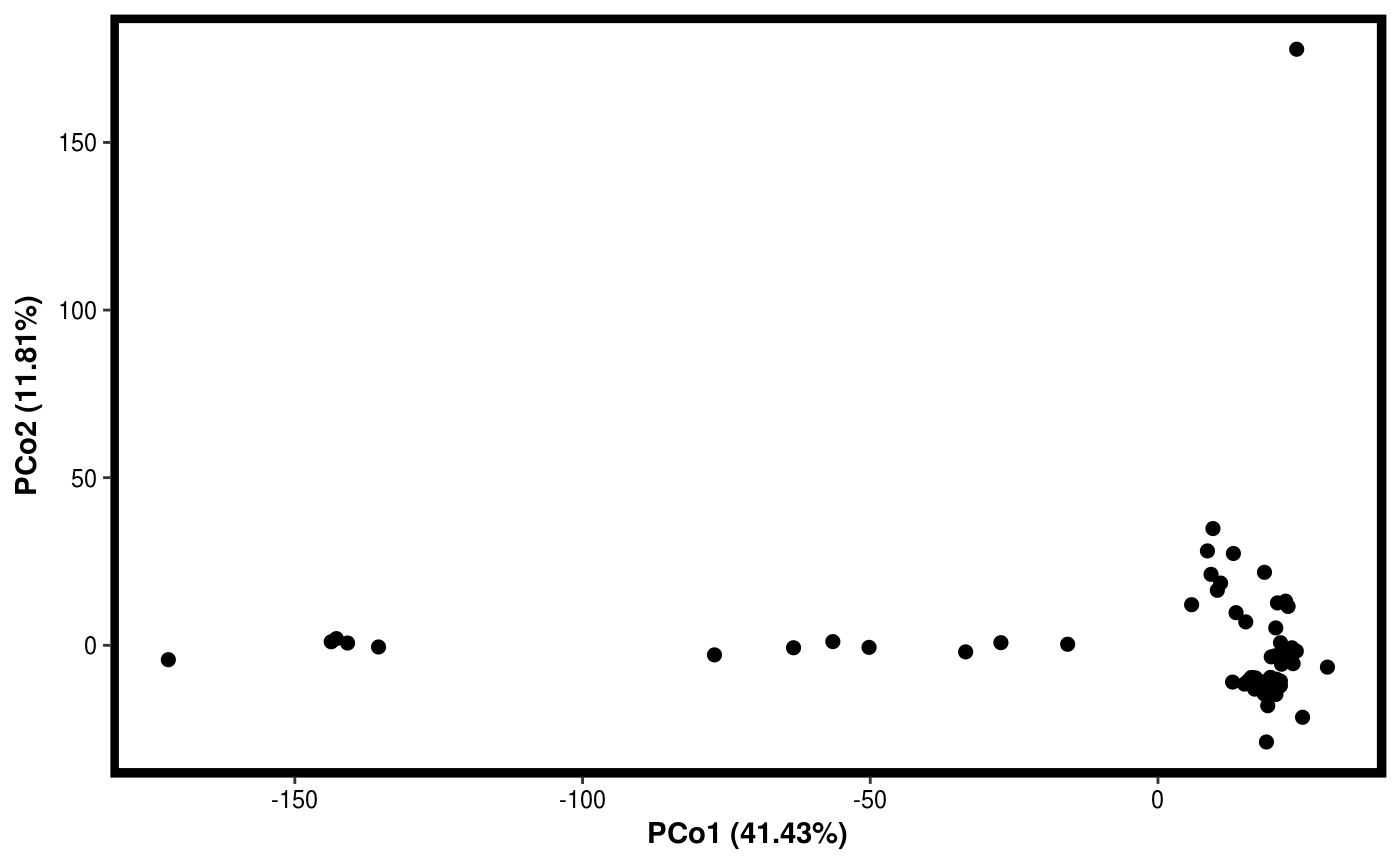
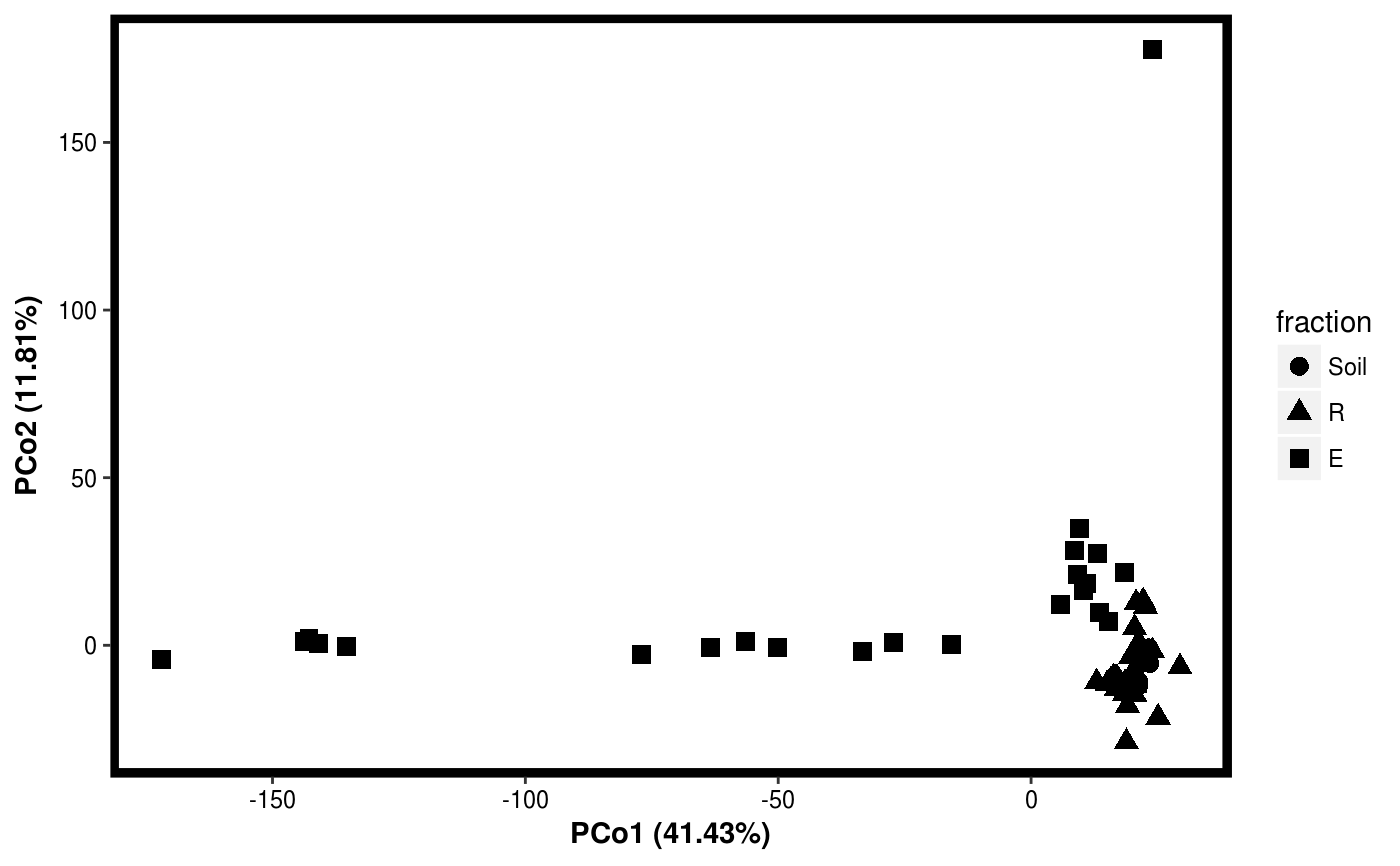
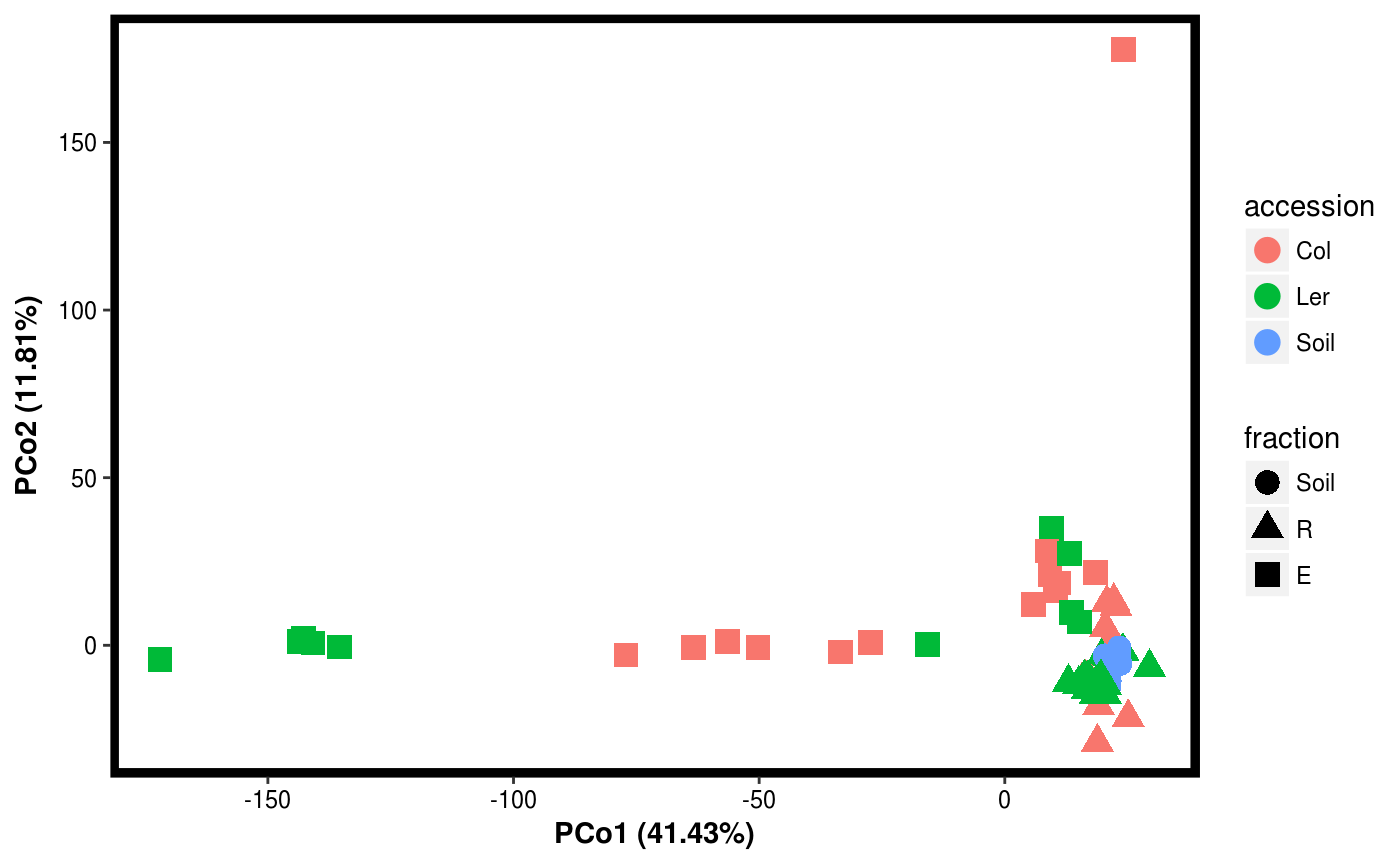
data(Rhizo) data(Rhizo.map) Dat <- create_dataset(Rhizo,Rhizo.map) # distfun <- function(x) vegan::vegdist(x,method="bray") #requires vegan package distfun <- dist Dat.pco <- PCO(Dat,dim=2,distfun=distfun) summary(Dat.pco)#> Principal Coordinate Analysis: #> 2 Components #> #> Component Var.explained Cumulative #> 1 PCo1 41.43 41.43 #> 2 PCo2 11.81 53.24plotgg(Dat.pco)