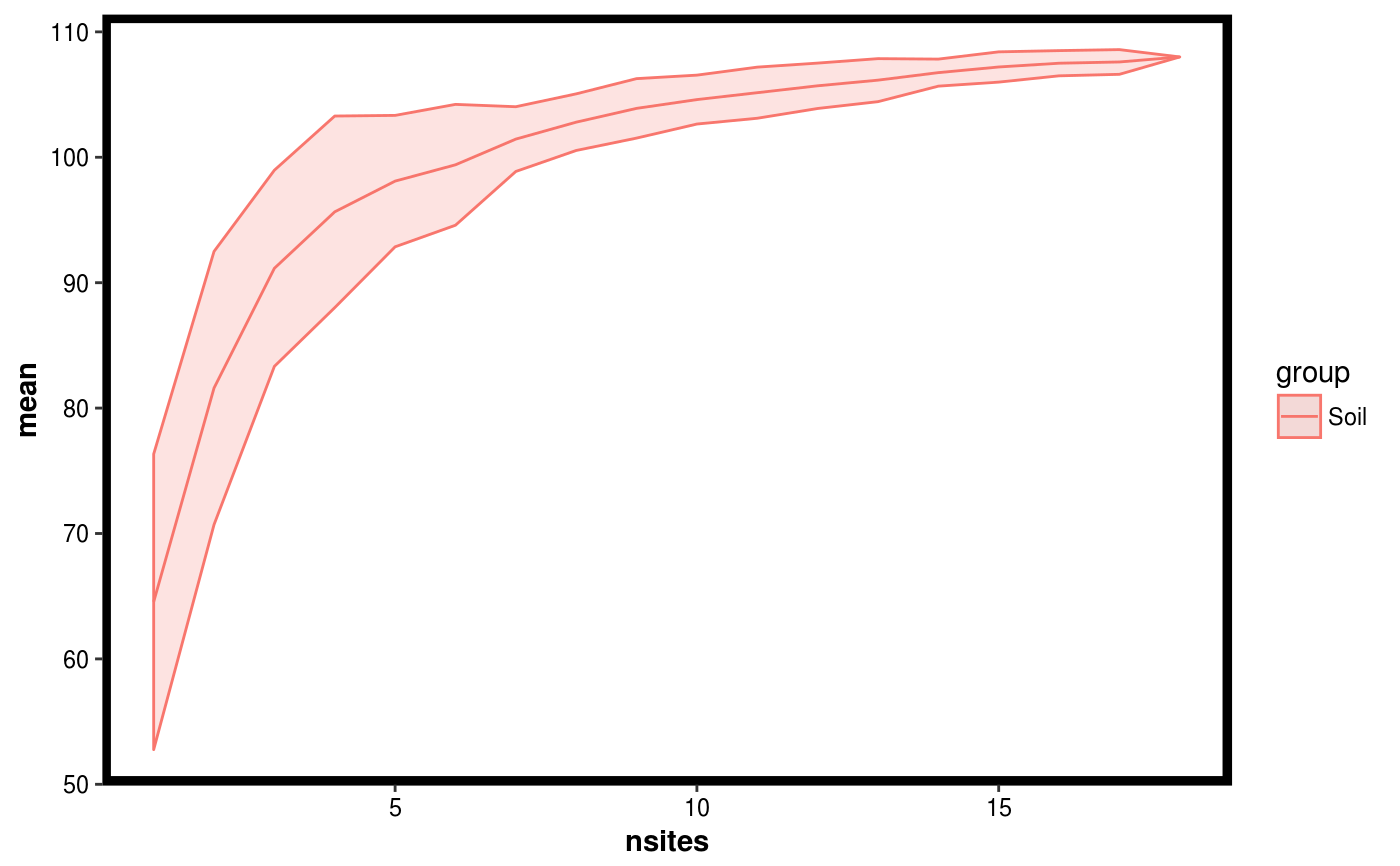
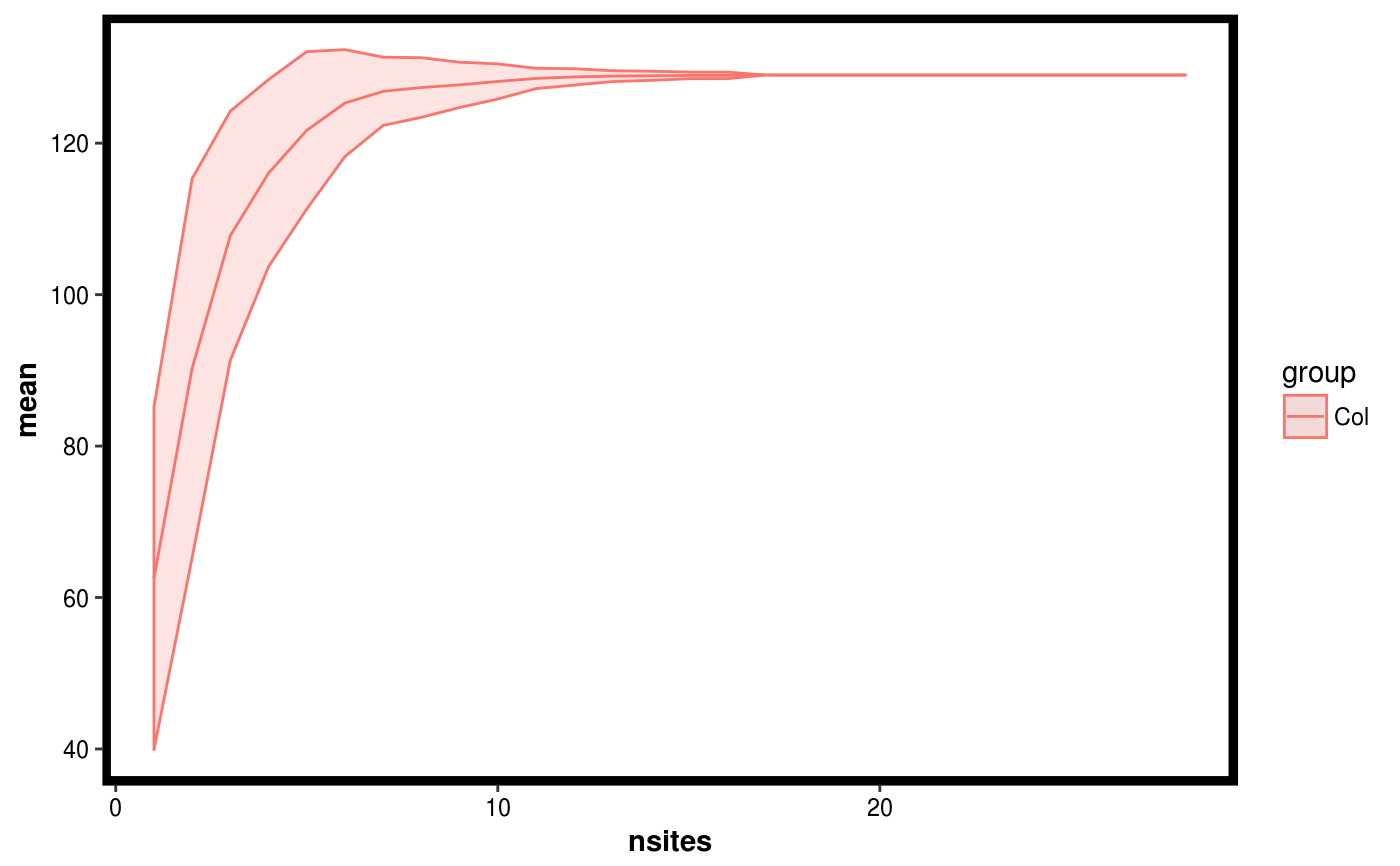
Calculates a diversity measure as a function of adding samples, and uses permutation to obtain a confidence interval.
site_diversity(Dat, factor, group, divfun = total_richness, nperm = 20)
Arguments
| Dat | A Dataset object |
|---|---|
| factor | String representing the name of the variable to be used for grouping samples. Must correspond to a header name in the Mat portion of the Dataset object. |
| group | Value of the variable defined in factor to extract. See examples. |
| divfun | Function that returns a diversity estimate given a matrix of samples. See total_richness and examples to see how to define your function. |
| nperm | Number of permutations to perform. |
Value
A data.frame of class site.diversity which contains the following variables:
- mean
Mean diversity value of all permutations
- sd
Standard deviation of the diversity estimates estimated from the permutations.
- nsites
Number of sites (ie. samples).
- group
Variable used for selecting the samples.
See also
Examples
data(Rhizo) data(Rhizo.map) data(Rhizo.tax) Dat <- create_dataset(Rhizo,Rhizo.map,Rhizo.tax) soil.sitediv <- site_diversity(Dat = Dat, factor = "accession", group = "Soil") col.sitediv <- site_diversity(Dat = Dat, factor = "accession", group = "Col") plotgg(soil.sitediv)plotgg(col.sitediv)# The following function requires the vegan package installed # and can be used with the site_diversity function. #divfun <- function(x){ # if(!is.null(ncol(x))) # x <- rowSums(x) # s <- vegan::diversity(x) # return(s) #}